Research Interests
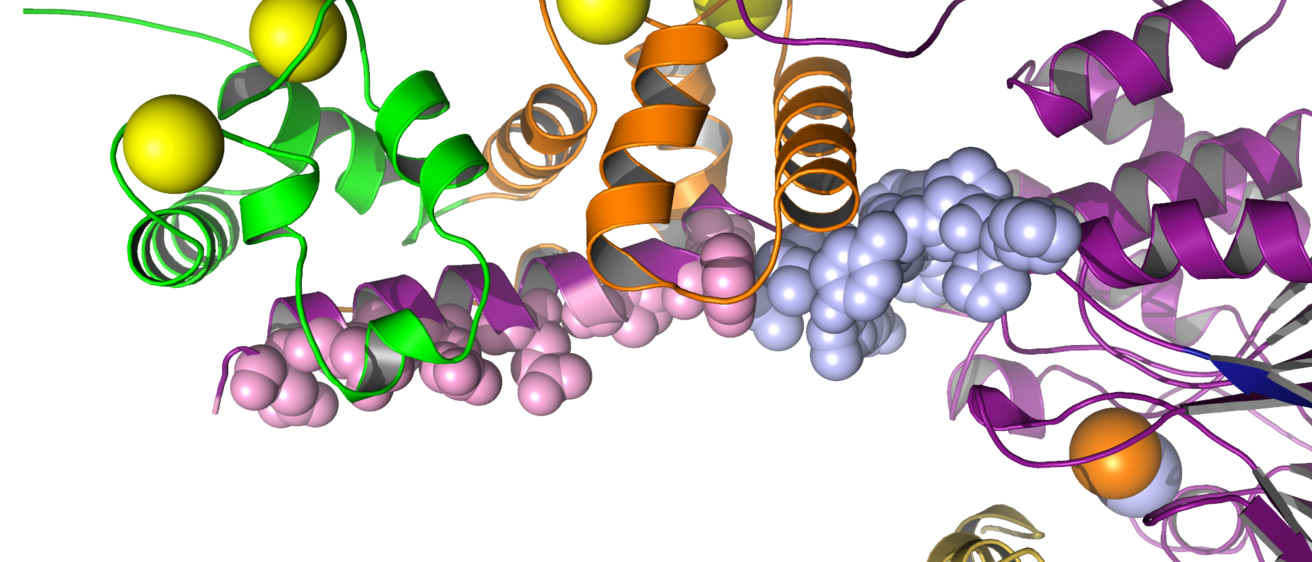
This laboratory probes the linkage between cooperative ligand binding, conformational change and enzyme activation by calmodulin, a regulatory calcium- binding protein. Calmodulin is the primary eukaryotic intracellular calcium receptor and serves as a second messenger to regulate cellular responses to transient calcium fluxes. It is clinically relevant for human physiology (for example, it is a target of anti-psychotic drugs) and is also found in plants and fungi. Cooperative binding of four calcium ions to calmodulin causes large conformational changes; these changes control the sites and extent of calmodulin activation of important cellular enzymes and structural proteins (see Figure).
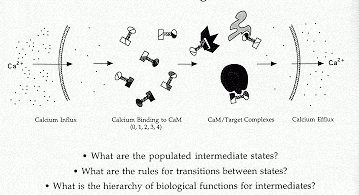
In order to determine the states that are functionally significant in this complex network of interactions, it is necessary to develop and apply new methods to directly determine energetic and structural properties of calcium binding. Quantitative proteolytic footprinting and applications of multi-dimensional heteronuclear NMR are capable of monitoring individual residues or bonds during a titration representative of a cellular influx of calcium. These studies have shown that the two domains of calmodulin interact in different ways as calcium fills the four sites of the protein.
Calcium-dependent structural rearrangements of calmodulin also are monitored by changes in the fluorescence, circular dichroism and hydrodynamic properties. These combined approaches are aimed at elucidating molecular mechanisms of cooperativity in calmodulin by determining the differences in intrinsic binding affinity at the four calcium-binding sites of calmodulin and the extent and nature of inter- and intra-domain cooperativity. To dissect these interactions, we are studying the isolated domains of calmodulin and many mutants shown to have phenotypic effects on complexes of calmodulin with its target enzymes.
Computational molecular modeling is used to visualize and calculate properties of these proteins and serves as a complement to the experimental studies of ligand- linked conformational change. The goal is to combine energetic and structural data to formulate models that will explain how synchronized changes in calcium levels modulate many diverse physiological processes.